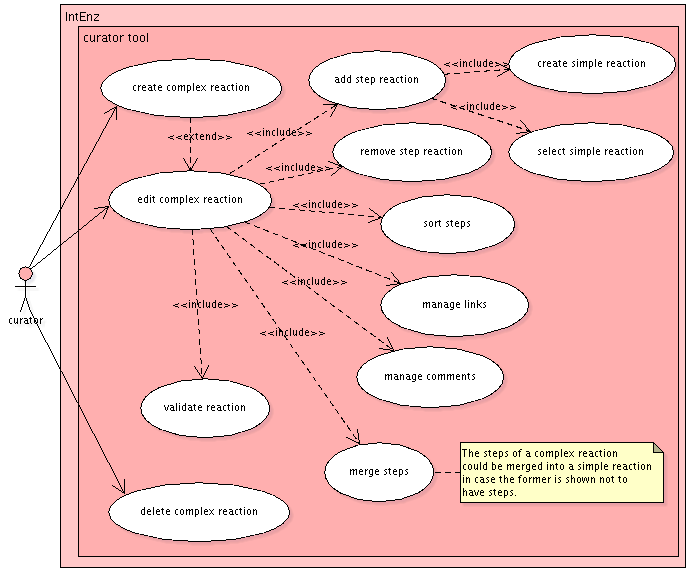
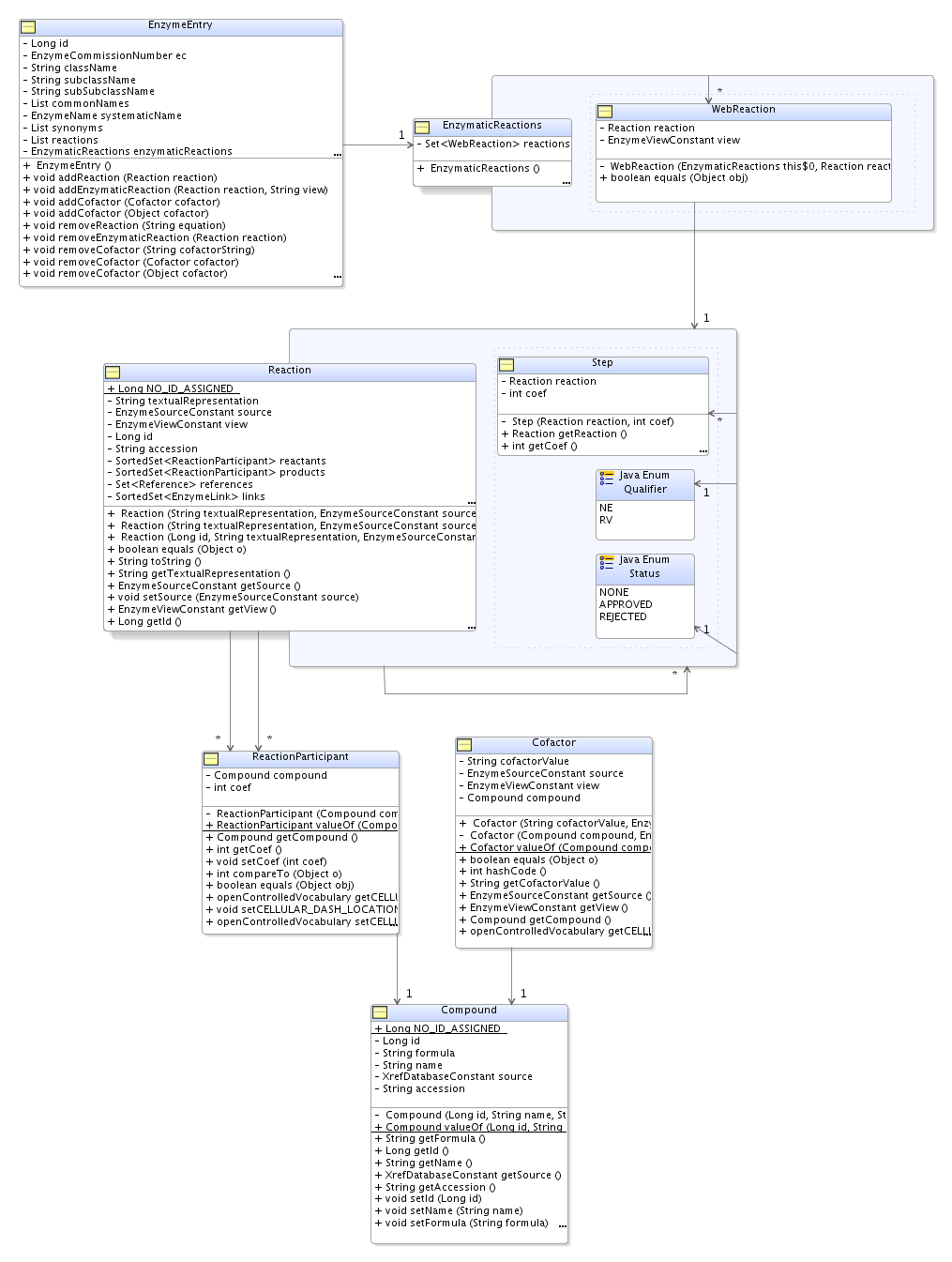
Current status and problems to solve
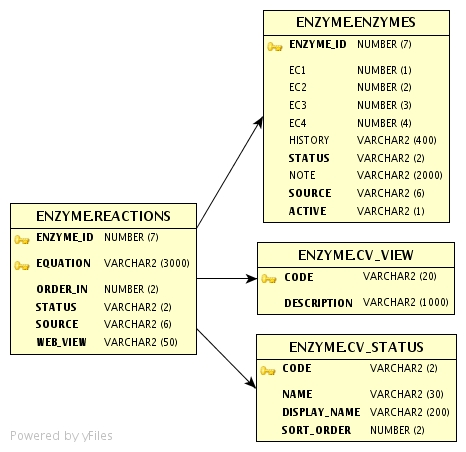
IntEnz includes one or more reactions per enzyme entry, as free
text. Those reactions have no unique identifiers, so if two
enzymes catalyze the same reaction, that means two rows in the
database containing the same equation string
(redundancy).
Reactions are ordered for an enzyme entry with multiple
reactions, but there is currently no way to distinguish reaction
steps and alternative reactions (ambiguity).
The string of characters which defines the equation has no link
to other databases for reactants and products
(isolation).
Enzyme entries have a unique EC number which can eventually
change due to transfer or splitting. Besides, the curator tool
architecture assigns them an internal unique identifier which
changes whenever the entry changes its status
(instability).
Goals
- Use a controlled vocabulary of small molecules available from ChEBI and map IntEnz reactions (reactants and products) and cofactors to it. To that end, the current reaction and cofactor text will be parsed, extracting the names of compounds which will be mapped to ChEBI.
- Stable and unique public identifiers for reactions. Once achieved, it will be feasible to develop stable public identifiers for the enzyme entries themselves.
Existing reaction databases and data models
Databases
- Reactome has
a different concept of reaction and enzyme compared to
IntEnz. It takes into account the compartment where
reactants and products are, and the species where
reactions are found. Besides, reactions are also considered
different because of their catalyst, which map to one
UniProt entry at most.
IntEnz, following NC-IUBMB design, considers a reaction as unique independently of the compartment, species or concrete catalyzing protein.- Database: Reactome uses the mySQL database and is pretty much tied to it
via different constraints. It uses a non-traditional approach to load its
schema from the database in that the schema is recreated from a 'schema_table'
in the database. The traditional approach used by most EBI databases is to design
tables and domain models that correspond, then use a mapping tool such as OJB,
Toplink or JDBC to map the tables to the domain model. This is not done in Reactome
this way because they anticipate many changes to their schema.
Advantages Disadvantages Generic schema format which could in theory map to any domain model. Non standard approach of database design - which means only a few specialised people could support it. -- Would require us to move to MySQL - which would cost a minimum 5 months full time development to move all our PL/SQL procedures developed for IntEnz. IntEnz does not using any generic mapping tools like Toplink or OJB so all the JDBC would need to be converted which would require a major revamp. -- Long term maintenance of this project would be at risk as mySQL is not officially supported by any DBA's in our group.
- Curator tool: Reactome has developed a standalone curator tool which uses
the JAVA swing framework. The tool is dependent on using the database schema
setup as described above. It is maintained by one developer based at the EBI.
The tool loads in a schema setup based on the database table 'schema_table'.
It then generates the domain model from this. It extracts an entry from the
database and loads it locally for the curator to use. This is kind of a like
a CVS facility. For any specialised treatment such as validation of the data you need to have
utility classes. The tool has no validation for reactions which is a component
we are desperately searching for.
Advantages Disadvantages Existing tool which is succesfully used in production by another group at the EBI. The tool is dependent on the database schema as described above. According to the developer if we deviate from using the generic database schema setup it would require a major revamp of the tool and in his opinion it would not be worth it. Using standard JAVA which is supported in our group. The validations which are currently done could cause a maintenance headache. Currently all validations are done via the STRUTS validation framework which ties in all the data neatly. The CVS facility is a great feature. Again the long term maintenance of this tool would be slightly worrying due to their non-standard approach to design. It is much easier to hire a developer who knows the STRUTS framework rather than a specific non-standard database tool. -- Using tool would require some major rework and it does not make sense in the light that IntEnz already has a specialised curator tool with all the enzyme functionality in it. The IntEnz curator tool is stable and has been in production for over a year, in addition the curators are finally happy with it.
- Database: Reactome uses the mySQL database and is pretty much tied to it
via different constraints. It uses a non-traditional approach to load its
schema from the database in that the schema is recreated from a 'schema_table'
in the database. The traditional approach used by most EBI databases is to design
tables and domain models that correspond, then use a mapping tool such as OJB,
Toplink or JDBC to map the tables to the domain model. This is not done in Reactome
this way because they anticipate many changes to their schema.
-
MACiE deals with enzymatic reaction mechanisms,
which means taking into account the protein structure (PDB
and CATH data). That is out of the scope of IntEnz.
As stated before, IntEnz reactions can't be different because of the catalyzing protein. MACiE reactions are. -
KEGG has public reaction ids. Reactions are
decomposed into reactants and products, with links to the
corresponding KEGG compounds. The database can be
searched - by reaction, compound or enzyme - using
webservices and a
jar file they provide. There is also a
user manual available.
Although the KEGG reaction database will provide a very good validation and cross-reference source we are unable to use it because it does not contain the strict CV formatting requirements which is a must for IntEnz.
However, we plan to use the KEGG reactions as an integral validation resource and also ease our cross mapping to reactions via the EC numbers.
Data Models
- SBML: the Systems
Biology Markup Language is becoming a standard format
for describing biochemical reaction networks. Based on XML
and other XML standards (MathML, CellML), it is used by an
increasing number of applications and databases
(BioModels and
Reactome among others).
SBML is not a database schema. Taken from the SBML FAQ:
There is no reason why SBML models could not be stored in a database, nor is there any reason why you could not use SBML as a schema. However, this was not the motivation for creating SBML. An SBML model is meant to encode a consistent view of knowledge of a biological system.
As BioPAX, SBML will be considered as as an import/export format for IntEnz reactions. To this end, there exist a jar file by JigCell to read/write SBML level 2. - BioPAX
is a collaborative effort to create a data exchange format
for biological pathway data using the OWL language.
Though it tries to represent the same type of data as
SBML, they differ in their views being BioPAX more
qualitative, cross-reference and ontology-oriented, and SBML
more quantitative and mathematics-oriented. Far from being
incompatible, they can be mixed in the same XML document.
BioPAX uses known standards, including pointers to GO, CML instances and SMILES format.
The Jastor project offers automatically generated APIs for several ontologies from their OWL files, among others a BioPAX API.
BioPAX will be a first choice import/export format for IntEnz reactions. In addition it is attractive to use some of the interfaces provided by BioPAX in our reactions java model as they meet some of the requirements we aim at (association of EC numbers to reactions and cross-referencing of reaction participants, for example).
The NC-IUBMB and ENZYME have strict requirements on how their data and therefore their controlled vocabularies are written. It is therefore not possible to use an external source as a controlled vocabulary as we would be unable to guarantee the formatting required by ENYZME and NC-IUBMB. For this reason we need to use ChEBI as our controlled vocabulary because
- it contains all the molecules needed by IntEnz,
- it is written in the XML format required by IntEnz and hence allows conversion to the other views, i.e. ENZYME and NC-IUBMB, and
- it decreases redundancy as we will not have to maintain our own controlled vocabulary.
Potential external users of enhanced IntEnz reactions
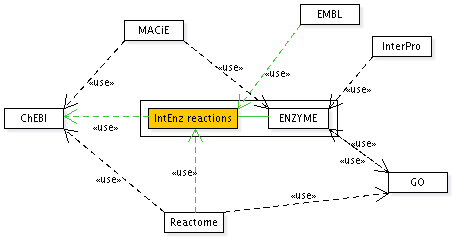
- EMBL:
the submissions to EMBL database include annotations with
EC numbers, which poses the same old problem of having
instable public identifiers. In that cases, EMBL could check
the EC no in IntEnz and keep a cross-reference to a
reaction id.
Another request from EMBL was having some kind of taxonomy information for the reaction. That is out of this specification's scope, but could be achieved by linking to Reactome of GO databases. - Reactome: Bernard de Bono has showed interest in linking to IntEnz reactions, as well as importing them into Reactome be it using web services or an XML interchange format (BioPAX).
- InterPro: metabolic pathways are described as sequences of InterPro entries, which map to EC numbers. Some of their entries have a reaction XML tag which contains a reaction as plain text. IntEnz reactions could be used by InterPro to link and update such information, in order to display reactions for InterPro entries. To that end, their curators would need to search the IntEnz reactions database.
Use-cases
Abstract reactions
An abstract reaction has no clear definition of the
biochemical process, the ambiguity coming from a broad range
of reactants and/or products. Thus, its participants cannot
be easily mapped to ChEBI. The only information for an
abstract reaction is a textual description.
This is the way in which every reaction is treated currently
in IntEnz.
Simple reactions
A simple reaction has no steps. It can be part of an overall (complex) reaction, i.e. be a step reaction.
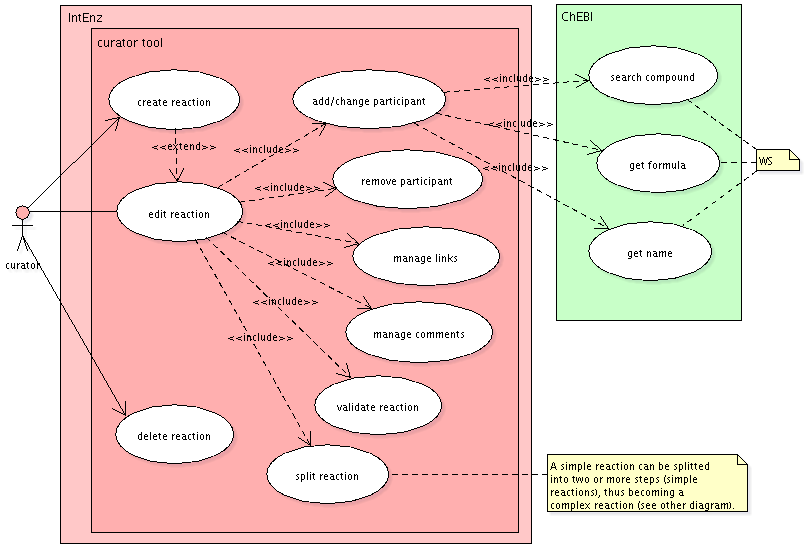
Deletion of reactions is actually a change of status (the data won't be removed from the database), much in the same way deletion of compounds is handled in ChEBI.
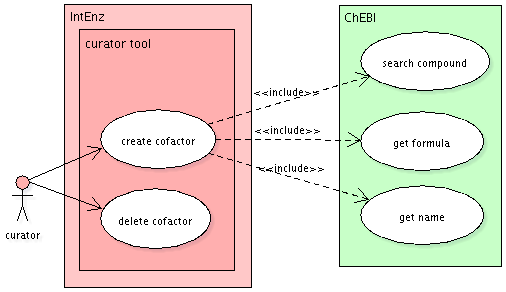
Cofactors
Though not being part of the reaction itsef, cofactors will be reimplemented to take advantage of the new mapping to ChEBI, favouring the reuse of data in IntEnz.
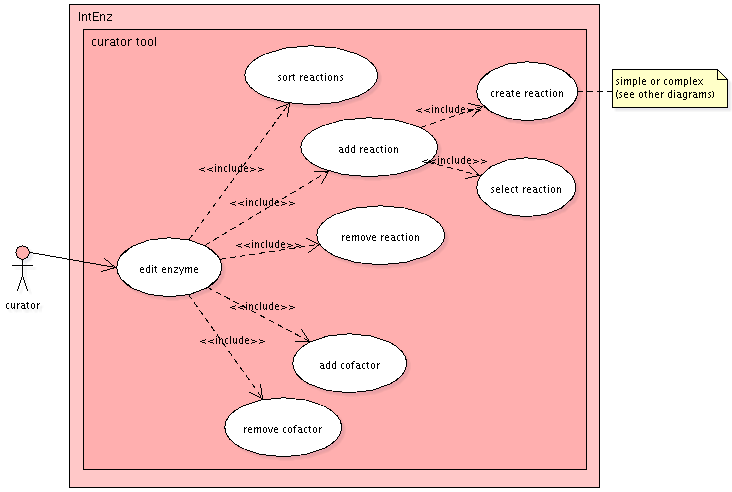
Enzyme entries
The assignment of reactions - simple or complex ones - to an
enzyme entry will allow curators to select from existing
reactions in the database, or create new ones if necessary.
Several reactions assigned to the same enzyme entry are
considered as alternative reactions. Sorting of alternative
reactions will be implemented as well.
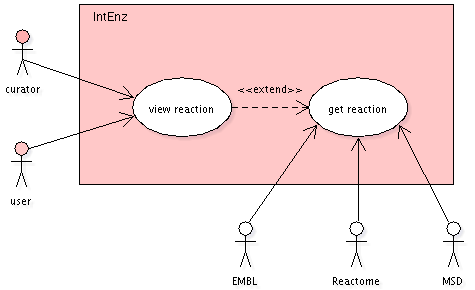
IntEnz reactions users
End users and curators will query IntEnz for reactions
(by EC number, reactant/product ChEBI id, reactant/product
name...) using a web interface.
External databases will update cross references to stable
public reaction id's, querying the database or - in the
future - using webservices.
Requirements
Dependencies on existing systems
The mapping of reactants, cofactors and products requires the use of the ChEBI database. It can be achieved in several ways:
- Acting as a direct client of the ChEBI database, using JDBC for example.
- Acting as a client of the ChEBI internal Web Services, using HTTP calls.
The retrieval of data from ChEBI would be only at curation time, to avoid overhead. So any needed compound information would be stored in the IntEnz DB and updated appropriately with every IntEnz release.
Platform requirements
IntEnz would need access to ChEBI, by adding the needed web services jar files and configuration files to the distribution.
Data fields required
An unbalanced ("abstract") reaction must have just a textual representation, like the current equation field.
A balanced ("chemical") reaction must meet the following requirements:
- Have one or more different reactants.
- Have one or more different products.
- No reactant may exist among products, and viceversa. (there are exceptions to this rule!, in which case the reaction could be handled as unbalanced).
- Reactants and products must map to other database(s) - namely ChEBI - from which data - name, formula ... - will be collected and updated. Besides, they will have a coefficient for stoichiometry purposes.
- The stoichiometry must be valid.
- The reaction must be the same independently of the participants names (synonyms) and ordering. (This is independent of the final representation, as some reactions will need some cosmetic reordering, to put water after any other participants, for example).
- Whenever a reactant/product is shown as an option - "an aldehyde or ketone", for instance - the reaction will split into alternative reactions.
A reaction will have four possible directions:
- "Left to right" (reactants -> products)
- "Right to left" (products -> reactants)
- Bidirectional
- Unknown directionality
In case it is known that one of the directions does never happen, its status - see below - will be set to obsolete and so will (automatically) bidi/unknown identifiers.
Reactions will have a status flag to avoid real deletions in the database and hence to enforce stable identifiers.
Reactions will be unique within the database, that is: one reaction could be assigned to more than one enzyme (reusable).
Reactions could have comments andlinks to other databases (KEGG reactions, MACiE). Links (to KEGG "IUBMB" reactions, for example) will be automated whenever possible.
Reactions will have qualifiers:
"Polymerization", "Class of reactions" and
"Chemically balanced"
("Spontaneous" has been discarded).
These qualifiers are not mutually exclusive, and should
be automated whenever possible, but still letting curators
to annotate the reactions.
Complex (overall) reactions will be made of two or
more single (step) reactions. Summing single
reactions will be possible, to yield a complex one.
Splitting a simple reaction into steps will convert it
into a complex one.
Validation will be applied to complex reactions as well as
their steps. Every step reaction will have a global
coefficient in order to balance apropriately the overall
reaction.
For now, just one level of complexity will be allowed, that
is a complex reaction of this type cannot be part (step)
of another complex reaction.
Step reactions will have a qualifier attached to it
which defines it as a "Primary" reaction or "Secondary"
reaction. (this was discarded, as it was really
meant to be applied to elementary reactions - see below -,
but even these cannot be qualified like this in an
absolute way)
Other type of complex (coupled) reactions will have no
specified steps, but will be the sum of two or more
simultaneous unordered elementary reactions, which
will act as reusable building blocks (for example,
hydrolysis of ATP).
These complex reactions without steps could act as steps
in an overall transformation.
Reactions which currently include an OR operator in its equation text ("an aldehyde or ketone", for example) would split into two alternative reactions for the enzyme entry.
Complex cofactors (text including the operators ";", "and", "or") will be splitand mapped to ChEBI. The OR operator could have several meanings (to be decided if one is enough).
Influence on existing systems
IntEnz database
The database schema should be modified as follows:
- The current REACTIONS table will become obsolete, as abstract reactions will also be stored in the new table...
- A new table INTENZ_REACTIONS as a hub for other tables containing detailed data. It will store essential data as well as columns to speed up reactions search - FINGERPRINT - and visualization - EQUATION.
- A new table REACTIONS_MAP to map enzyme ids to reaction ids, as well as store the ordering of alternative reactions and web view for complex reactions (should their steps be shown?).
- A new table COMPLEX_REACTIONS will store a mapping for overall reactions to their steps, as well as their ordering and coefficient (if needed to balance the overall equation).
- A new table REACTION_LINKS for any cross references to KEGG reactions, UM-BDB...
- A new table REACTION_CITATIONS for any bibliographic references to the reaction.
- A new table CV_REACTION_QUALIFIERS.
- A new table CV_REACTION_ROLES will store the two possible roles in a reaction.
- A new table REACTION_PARTICIPANTS will link reactions to their reactants and products.
- A new table CV_REACTION_PARTICIPANT_ROLE.
- A new table COMPOUND_DATA will store data retrieved from ChEBI.
- The COFACTORS table will have a new column COMPOUND_ID, which will be primary key instead of COFACTOR_TEXT. It will also have two more columns: OPERATOR and OP_GRP, to handle complex cofactor texts.
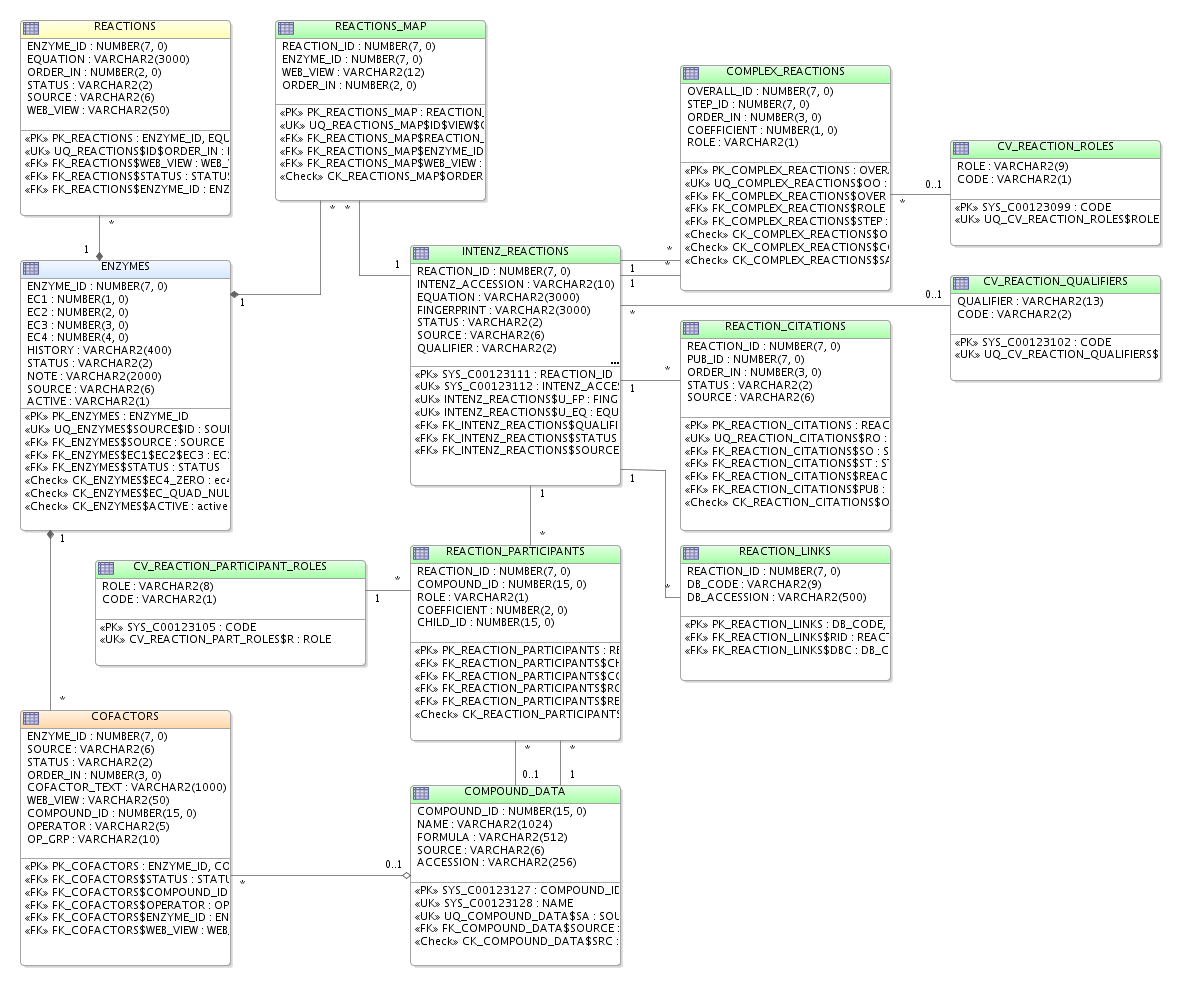
There is an example available (outdated, as of 2006-10-31).
As part of the future data integrity checker:
- ChEBI id's used by IntEnz should be available in ChEBI, and should not be child compounds (these will be substituted by their parents in case of merging, during routine automated ChEBI updates ). Whenever there is a chenge in ChEBI which propagates to IntEnz, both ChEBI and IntEnz curators should be warned to be aware of possible side effects (on enzyme.dat, for example).
- the stoichiometry of every reaction should be validated, not only at curation time but also after any ChEBI updates which could break the balance.
IntEnz curator tool
The classes ReactionDTO and CofactorDTO should be modified according to Reaction and Cofactor.
A validation for stoichiometry - possibly based on the compounds formulae - should be implemented in the biobabel package and used by the ReactionDTO class.
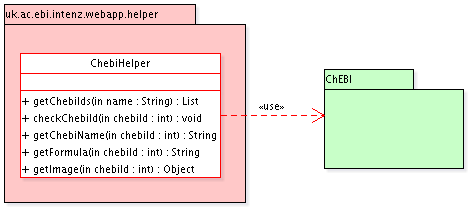
A new class ChebiHelper
should provide methods to use it as a ChEBI webservices
client, retrieving information on the reaction participants:
recommended name, formula, structure (image)...
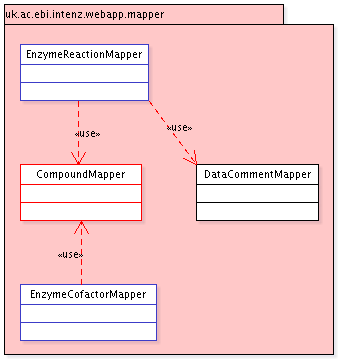
A new class CompoundMapper would be
responsible for retrieving compound data from the IntEnz
database (data already imported from ChEBI). It should be
used by EnzymeReactionMapper and
EnzymeCofactorMapper.
The web interface should be extended to create and edit reactions (see a preliminary model. Instead of a plain text field, buttons will be implemented to add alternative reactions, add steps or edit an existing reaction; a reaction editor will show up, including:
- Lists of reactants and products
- Text fields and buttons to populate the mentioned lists with their content
- Fields to manage links to other databases: GO, Reactome, KEGG...
- Fields for comments on the reaction
- While the curator is selecting a compound.
- After the curator has submitted a compound name/id.
Editing a reaction shared by two or more enzymes should display a warning message to avoid undesired changes.
A post-condition should be added to the edition of new reactions: it must not be already present in IntEnz, otherwise the existing reaction will be used. The check would need a fingerprint of the reaction generated from its participants.
Conclusions
- Database: The database schema designed above has been viewed and corrected
by various database specialists at the EBI. In addition the database has been
designed with similar systems in mind such as Reactome. It was not possible
to exactly adopt the Reactome database schema as it was far more complex than
required by IntEnz. However the essential items such as Inputs and Outputs were
adopted.
There were certain constraints imposed on the Reaction schema due to our engagement with the NC-IUBMB data schema. For example, we still need to handle unbalanced reactions which are unlikely to be dealt with by NC-IUBMB in the near future. Also the cloning of enzymes means that we are forced to keep the enzymes and reactions in the same schema. - Domain Model: The Java Domain Model will be implemented using interfaces from the BioPAX project. The advantage is that we are standard compliant and because it's just an interface (it could always be easily removed if need be) we are not creating an unnecessary dependency. The decision was taken because the API resembled what we are trying to achieve with our reactions. Unfortunately there is no validation framework implemented in any of the domain models we looked at. SBML is another possible output format but it did not seem suitable to use it inhouse.
- Curator tool: At this point in time there seems no clear advantage to use the Reactome Curator tool over our existing IntEnz tool. The IntEnz curator tool is a standard based system using the STRUTS web framework, Unit Of Work design pattern, STRUTS validation framework, TILES framework, JSTL tag libraries and STRUTS tag libraries. In addition the extention of this tool to accomodate Reactions would be minimal. Another advantage is that we have spent months ironing out the bugs and preferences that SIB and EBI curators had for the tool. It is now in an acceptable state. Introducing a new tool which would require the same amount of effort and bug fixes could cause major havoc to the colloborative spirit of this project.
Timetable
We estimate the project will take approximately 9 months. We have spent about 2 months gathering requirements and writing this specification, hence we estimate in another 7 months we expect the project to be complete. See below for the project deliverables:
| Priority | Description | Developer | Estimate |
| 1. | Reaction database design and modelling. | RA and PdM | 10 d |
| 2. | Reaction database implementation. | RA | 20 |
| 3. | Reaction mapping to ChEBI and ChEBI import. | RA | 45 |
| 4. | Reaction validator. | RA | 20 |
| 5. | Curator tool implementation of Reaction database. | RA | 40 |
| 6. | Public tool implementation of Reaction database. | RA | 20 |